library(tidyverse)
source("R/set_ggplot_theme.R")
library(broom)
library(Rmisc)
library(car)
library(lme4)
library(lmerTest)
library(nlme)
library(VCA)
library(afex)QK Box 10.6
Caballes et al. (2016) examined the effects of maternal nutrition (three treatments: starved or fed one of two coral genera: Acropora or Porites) on the larval biology of crown-of-thorns seastars. There were three female seastars nested within each treatment, 50 larvae reared from each female were placed into each of three glass culture jars and the lengths of ten larvae from each jar after four days were measured after 4 days. This fully balanced design has maternal nutrition as a fixed factor with three random levels of nesting: females within nutrition treatment, jars within females and individual larvae within jars.

The paper is here
Caballes, C. F., Pratchett, M. S., Kerr, A. M. & Rivera-Posada, J. A. (2016). The role of maternal nutrition on oocyte size and quality, with respect to early larval development in the coral-eating starfish, Acanthaster planci. PLoS One, 11, e0158007.
Preliminaries
Packages:
Data:
caballes_length <- read.csv("data/caballes_length.csv")set_sum_contrasts()Make female and jar factors
caballes_length$female <- factor(caballes_length$female)
caballes_length$jar <- factor(caballes_length$jar)Reorder nutrition treatments so starved is first for default lm contrasts
caballes_length$diet <- factor(caballes_length$diet, levels = c("starved","aa","pr"))
levels(caballes_length$diet)[1] "starved" "aa" "pr" Visualize data
caballes_length %>%
ggplot(aes(jar, length, color = female)) +
geom_point(alpha = 0.5) +
facet_wrap(~ diet, scales = "free_x")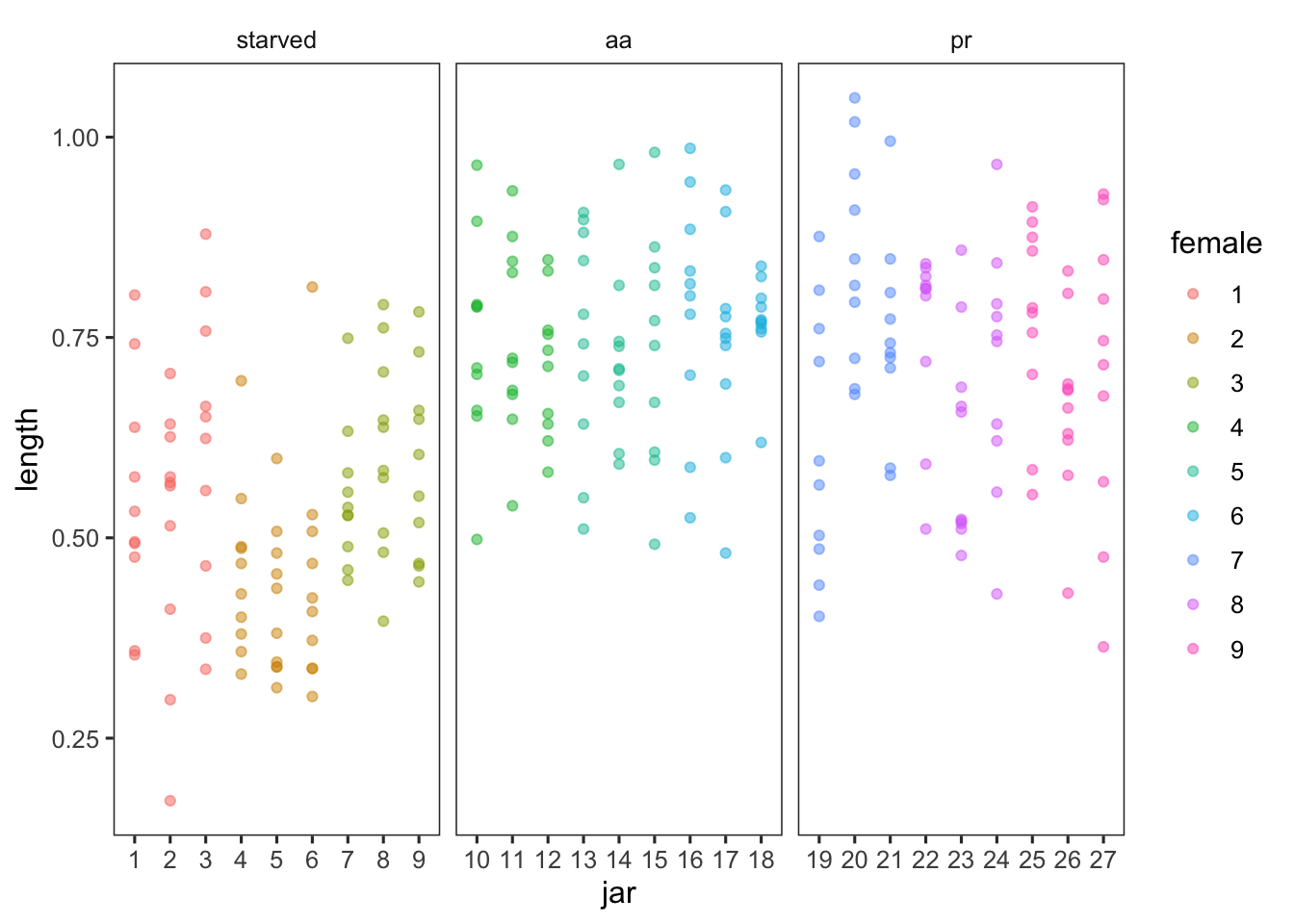
Run model
Note: can’t get the aov commands to work for 3 level nested design
caballes.aov <- aov(length~diet+Error(female/jar), caballes_length)Fit as lm using OLS estimation
caballes.lm <- lm(length ~ diet/female/jar, caballes_length)
anova(caballes.lm)Analysis of Variance Table
Response: length
Df Sum Sq Mean Sq F value Pr(>F)
diet 2 2.5334 1.26668 72.1624 < 2.2e-16 ***
diet:female 6 0.3735 0.06224 3.5460 0.002199 **
diet:female:jar 18 0.5265 0.02925 1.6664 0.045989 *
Residuals 243 4.2654 0.01755
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Check diagnostics and lm output (not shown):
plot(caballes.lm)
summary(caballes.lm)Get F and P values using correct denominators
#Diet F
f <- 1.26668/0.06224
pf(f, df1 = 2, df2 = 6, lower.tail = FALSE)[1] 0.002120396#Females F
f <- 0.06224/0.02925
pf(f, df1 = 6, df2 = 18, lower.tail = FALSE)[1] 0.100229#Jars F
f <- 0.02925/0.01755
pf(f, df1 = 18, df2 = 243, lower.tail = FALSE)[1] 0.04594475Variance components from VCA
caballes.vca <- anovaMM(length ~ diet/(female)/(jar), caballes_length)
caballes.vca
VCAinference(caballes.vca, alpha = 0.05, VarVC = TRUE, ci.method = "satterthwaite")Fit mixed effects model using REML/ML
caballes.lmer <- lmer(length ~ diet + (1|female/jar), caballes_length)
summary(caballes.lmer)Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: length ~ diet + (1 | female/jar)
Data: caballes_length
REML criterion at convergence: -289.2
Scaled residuals:
Min 1Q Median 3Q Max
-2.69014 -0.69659 0.01388 0.64143 2.59374
Random effects:
Groups Name Variance Std.Dev.
jar:female (Intercept) 0.00117 0.03420
female (Intercept) 0.00110 0.03316
Residual 0.01755 0.13249
Number of obs: 270, groups: jar:female, 27; female, 9
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 0.66279 0.01518 5.99986 43.652 9.68e-09 ***
diet1 -0.13588 0.02147 5.99986 -6.328 0.000728 ***
diet2 0.08301 0.02147 5.99986 3.866 0.008306 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) diet1
diet1 0.000
diet2 0.000 -0.500Get F-ratio for diet test using lmerTest
anova(caballes.lmer, ddf = "Kenward-Roger")Type III Analysis of Variance Table with Kenward-Roger's method
Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
diet 0.71442 0.35721 2 6 20.35 0.002121 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1CI on variance components (remembering to square CIs from lmer which are in SD units)
caballes.ci <- confint.merMod(caballes.lmer)
caballes.vc <- (caballes.ci)^2
print(caballes.vc) 2.5 % 97.5 %
.sig01 0.000000000 0.004051007
.sig02 0.000000000 0.003177589
.sigma 0.014769187 0.021083239
(Intercept) 0.404053917 0.475997080
diet1 0.030364676 0.009506432
diet2 0.001992217 0.014735036Simplify dataset by averaging across larvae within a jar
d <- caballes_length %>%
group_by(diet, female, jar) %>%
dplyr::summarise(mean = mean(length),
sd = sd(length),
n = n(),
se = sd / sqrt(n)) %>%
ungroup()
d %>%
ggplot(aes(female, mean)) +
geom_point(alpha = 0.5) +
facet_wrap(~ diet, scales = "free_x") +
labs(y = "Mean length (mm)")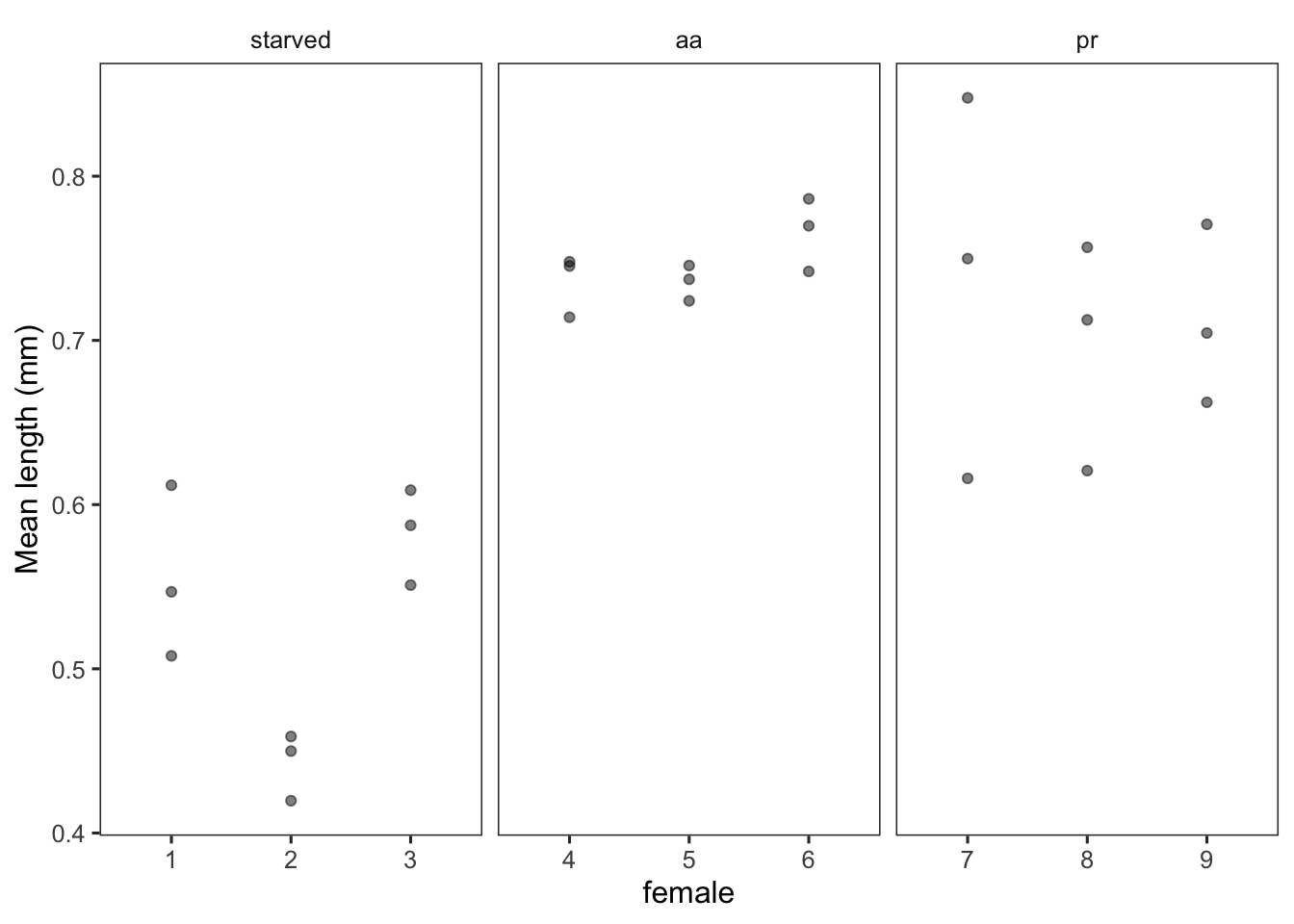
Fit nested ANOVA with OLS
Female is nested within treatment:
d_aov <- aov(mean~diet+Error(female), d)
summary(d_aov)
Error: female
Df Sum Sq Mean Sq F value Pr(>F)
diet 2 0.25334 0.12667 20.35 0.00212 **
Residuals 6 0.03735 0.00622
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Error: Within
Df Sum Sq Mean Sq F value Pr(>F)
Residuals 18 0.05265 0.002925 m1 <- lm(mean ~ diet / female, d)
anova(m1)Analysis of Variance Table
Response: mean
Df Sum Sq Mean Sq F value Pr(>F)
diet 2 0.253336 0.126668 43.3035 1.323e-07 ***
diet:female 6 0.037346 0.006224 2.1279 0.1002
Residuals 18 0.052652 0.002925
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Get F and P values using correct denominators. Note that I’m using broom:tidy to tidy the anova output, and lead() to calculate the new_F and new_P. This avoids extracting statistic (F-value) and df into new objects, or hard-coding the calculations.
tidy(anova(m1)) %>%
mutate(new_F = meansq / lead(meansq),
new_P = pf(new_F, df1 = df, df2 = lead(df), lower.tail = FALSE)) %>%
kable(digits = 3)| term | df | sumsq | meansq | statistic | p.value | new_F | new_P |
|---|---|---|---|---|---|---|---|
| diet | 2 | 0.253 | 0.127 | 43.303 | 0.0 | 20.351 | 0.002 |
| diet:female | 6 | 0.037 | 0.006 | 2.128 | 0.1 | 2.128 | 0.100 |
| Residuals | 18 | 0.053 | 0.003 | NA | NA | NA | NA |
Fit mixed effects model using REML/ML
m2 <- lmer(mean ~ diet + (1|female), d)
summary(m2)Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: mean ~ diet + (1 | female)
Data: d
REML criterion at convergence: -58.6
Scaled residuals:
Min 1Q Median 3Q Max
-2.05994 -0.46141 0.08908 0.48149 2.22410
Random effects:
Groups Name Variance Std.Dev.
female (Intercept) 0.001100 0.03316
Residual 0.002925 0.05408
Number of obs: 27, groups: female, 9
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 0.66279 0.01518 6.00000 43.653 9.68e-09 ***
diet1 -0.13588 0.02147 6.00000 -6.328 0.000728 ***
diet2 0.08301 0.02147 6.00000 3.866 0.008305 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) diet1
diet1 0.000
diet2 0.000 -0.500anova(m2, ddf = "Kenward-Roger")Type III Analysis of Variance Table with Kenward-Roger's method
Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
diet 0.11906 0.059528 2 6 20.35 0.002121 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1